Search
Search Functions
There are a number of functions in the Search pull-down menu.

There are three general types of searches available here: MegaSearch, BLAST, and Standard searches. Each of these are described on the following pages.
MegaSearch
MegaSearch functions as a broad-functioning search tool to query a variety of types of information.
Other information types may be added in the future. Many of these search functions can also be found in individual links in the Search menu, but MegaSearch offers additional ways to subset data. For example, Gene/Transcript searches can be subset by annotations from BLAST, KEGG, InterPro, GO Term, and GenBank.
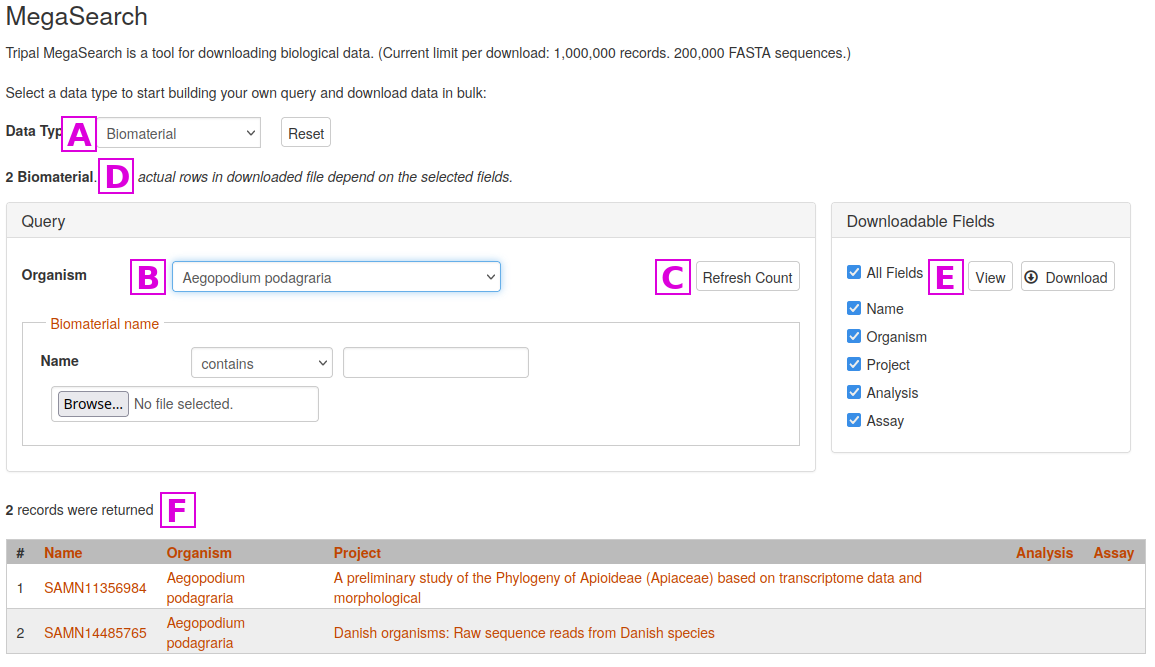
To perform a search with MegaSearch, first select the desired Data Type [A]
Then enter one or more search critera. In this example, we are just specifying the Organism [B]
Results do not appear automatically in MegaSearch. Once criteria are specified, click on the Refresh Count button [C], and the count of the number of results will be updated at [D]
To view the results, click on the View or Download buttons [E]. Here you can also specify which columns appear in the output. If you select the View button, results will appear at the bottom of the screen [F]
BLAST
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The CarrotOmics BLAST+ tool provides functionality similar to the well known BLAST tool at NCBI as implemented here by the Tripal BLAST module. CarrotOmics provides links to blastn, blastx, tblastn, and blastp analyses.
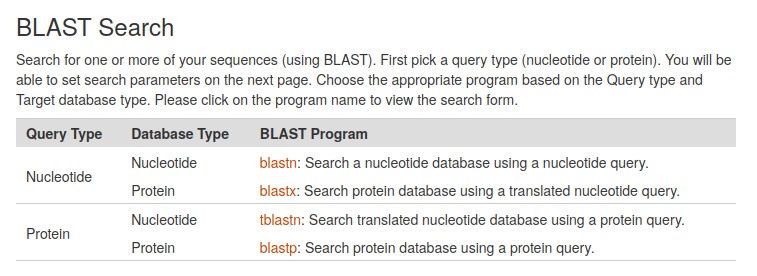
Once the BLAST program of interest has been selected, a query sequence can be entered into a search box, or a fastA file can be uploaded. Users can also select a blast database and specify advanced BLAST algorithm parameters.
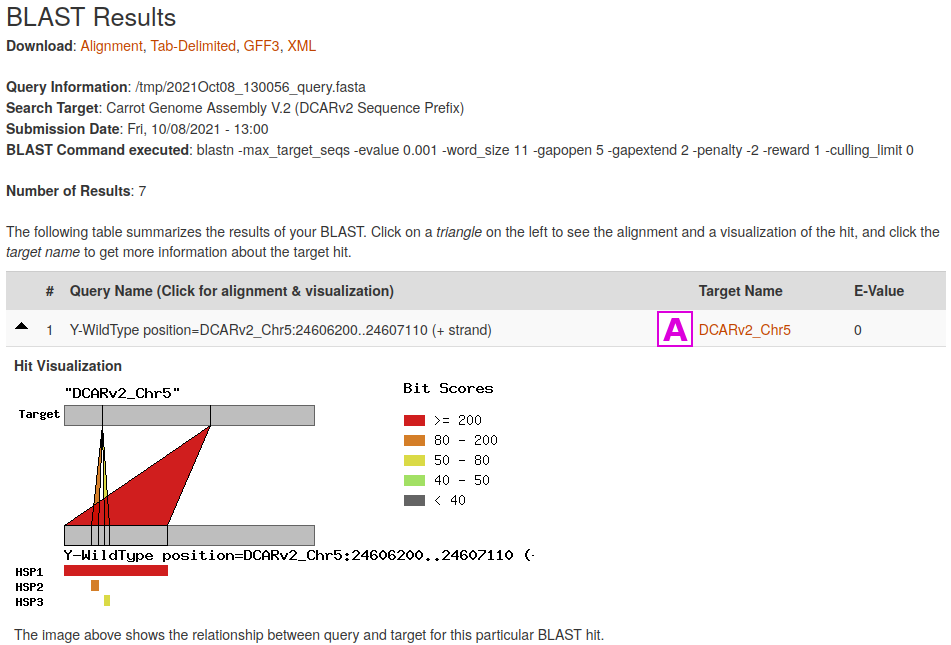
The Tripal BLAST module displays results in an interactive interface, where alignment to the target sequence can be viewed. Features that are in the CarrotOmics JBrowse will have clickable links to their appropriate entries [A].
Standard Searches
The remaining searches listed in the top-level Search menu all utilize similar interfaces.
Germplasm Search can be used to find accessions by name, keywords such as a trait or germplasm collection, taxonomy, country of origin, and cultivar pedigree. Additionally, those accessions with images can be searched by name, plant organ, and year the image was collected.
The Gene and Transcript Search can be used to identify genome features including the NCBI Daucus Annotation release 100, and reference genome gene prediction. Searches can also be completed in specific physical regions, by the name of the gene or transcript, or keywords related to a gene.
Linkage maps uploaded to CarrotOmics are searchable via the Map Search function. Maps can be subset to include only wild carrot (Daucus carota subsp. carota) or cultivated carrot (Daucus carota subsp. sativus). Results of map searches are organized into a table with a link to the map, descriptions of the mapping population including parental accessions, mapping population size and generation. A related search function is Marker Search. Markers can be found based on (sub)species, marker type, mapping population, linkage group, and if available the trait that the marker is associated with from the mapping study or association analysis. “Marker Search” may also be used to identify markers near regions of interest for QTL confirmation of selecting markers to test in genomic-assisted selection.
QTL and MTL Search, Qualitative Trait Search, and Quantitative Trait Search are also search functions related to genetic mapping. QTLs and MTLs can be searched by trait name. Specific traits can also be searched in the “Qualitative Trait Search”, and “Quantitative Trait Search”, differentiated by the traits being categorically or continuously scored, respectively. Searches are guided by drop down menus containing trait names or specific phenotypes scores.
CarrotOmics currently stores 40,497 publication references. These publications represent a subset of the scientific literature of particular interest to Apiaceae researchers, based on relevant keywords and manual curation. Searches for specific topics can be completed on the Publication Search page. Multiple keywords from the abstract, journal name, authors, or title can be used to search for specific Apiaceae topics. For example, searching ‘Carrot’ and ‘Cow’ in the title results in two publications that studied the effect of carrot and/or β-carotene added to the diet of cows.
Published primers can be searched in the Primer Search link from previous analyses, or can be searched either by name or by primer sequence, in either forward or reverse complement orientation, using degenerate base codes if desired. For example, searching with the identifier ‘MYB’ results in four primers related to MYB transcription factors. Finally, specific downloadable data and experimental information can be searched in the Project, Analysis, or Assay Search link.