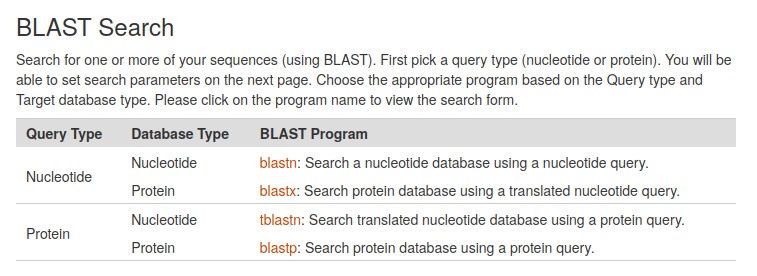
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The CarrotOmics BLAST+ tool provides functionality similar to the well known BLAST tool at NCBI as implemented here by the Tripal BLAST module. CarrotOmics provides links to blastn, blastx, tblastn, and blastp analyses.
Once the BLAST program of interest has been selected, a query sequence can be entered into a search box, or a fastA file can be uploaded. Users can also select a blast database and specify advanced BLAST algorithm parameters.
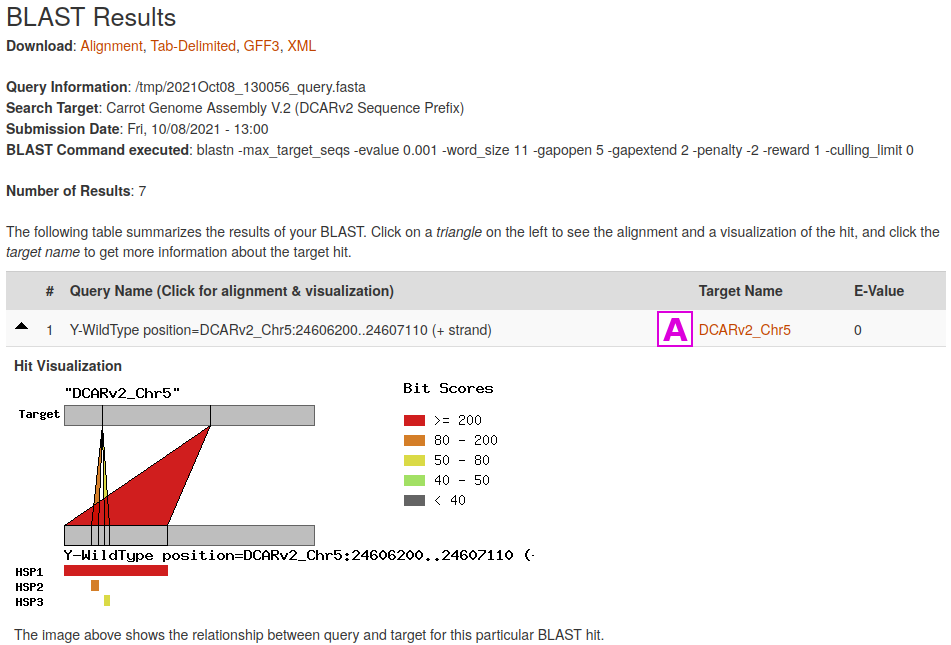
The Tripal BLAST module displays results in an interactive interface, where alignment to the target sequence can be viewed. Features that are in the CarrotOmics JBrowse will have clickable links to their appropriate entries [A].