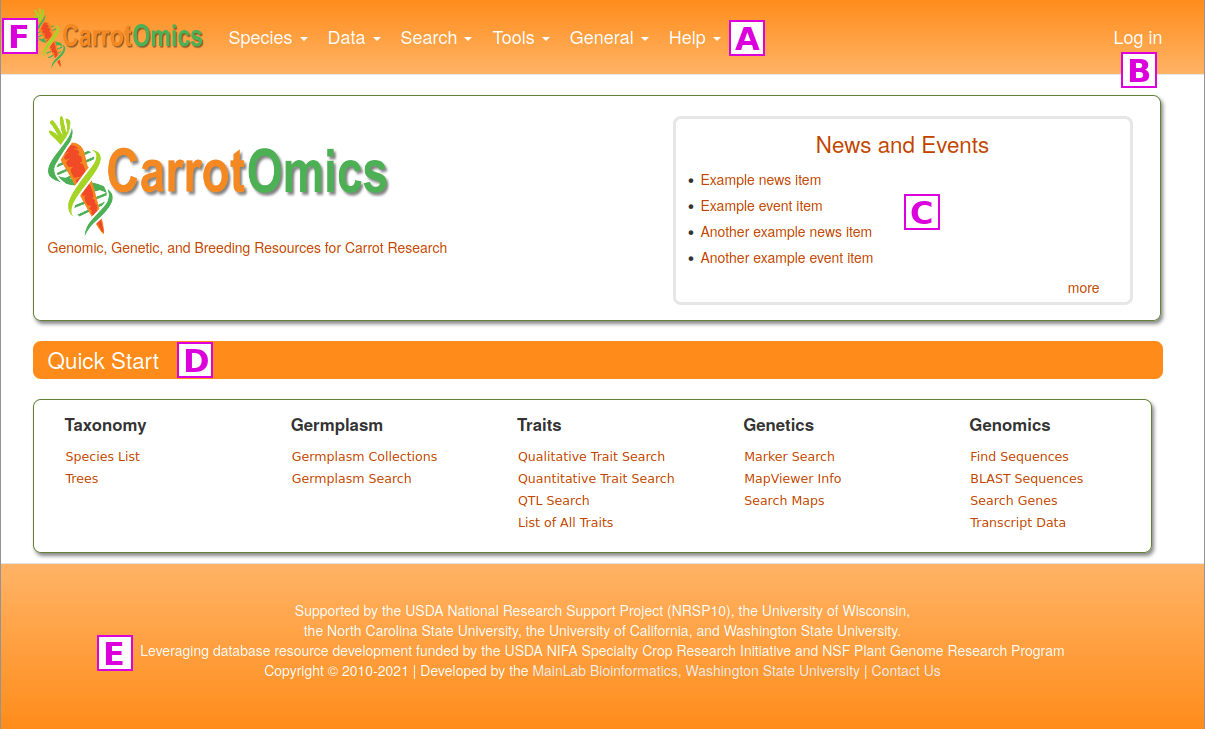
The CarrotOmics homepage is divided into a few key areas to help facilitate navigation.
[A] A traditional pull-down menu bar in the header provides access to all of the site's functionality.
[B] A link to log in to CarrotOmics. This is only used by administrators for uploading new data and performing maintenance.
[C] The center region of the homepage contains a News and Events section which has news about CarrotOmics as well as from the community.
[D] Commonly used functions are provided in the Quick Start section of the home page for easy access.
[E] Funding and technical support information.
[F] You can return to the home page at any time by clicking on the CarrotOmics logo in the upper left corner of the current page.
The items under the Search and Tools menus are discussed in other tutorials, but here we want to highlight the Data, General and Help menus. Under the Data menu [G] you can see a data overview, learn how to submit data, information about the types of genetic markers in CarrotOmics, and transcript and expression data. A summary overview of all data in CarrotOmics which can be broken down by species and/or data type is found on the Data Overview page. Under Data Submission, there are details about submitting data to CarrotOmics. CarrotOmics will accept published data and we highly recommend contacting us before starting to fill in the data templates. There are two links to the contact form on this page.
On the Marker Types page, there is a table of the marker types that are in CarrotOmics. The markers have been generally grouped by the same technologies and we have provided a link to a reference that describes the marker technology.
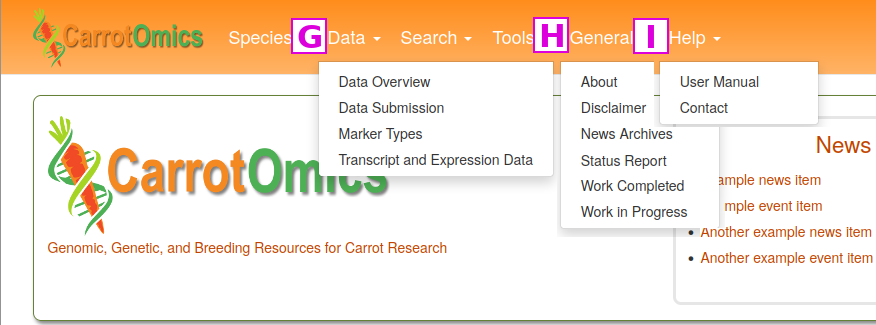
The General menu [H] contains links to a variety of different items.
The Help menu [I] has a link to this CarrotOmics User Manual and video tutorials. There is also a Contact Us link that opens a fillable web form. We appreciate input on the website as well as reports of website bugs and data errors.
